LOCUST
LOCUST (LOcus CUstom Sequence Typer) is a custom sequence locus typer tool for classifying microbial genomes. It provides a fully automated opportunity to customize the classification of genome-wide nucleotide variant data most relevant to biological research.
Key Features
- Ability to type bacterial genomes using any user-supplied DNA sequence.
- Automated processing from genome sequence download to genotypic classification and phylogenetic inferences.
- User customization enabling the processing of existing or generation of novel, classification schemes.
- Genomic assertions based on genotypic classifications.
Sample Output
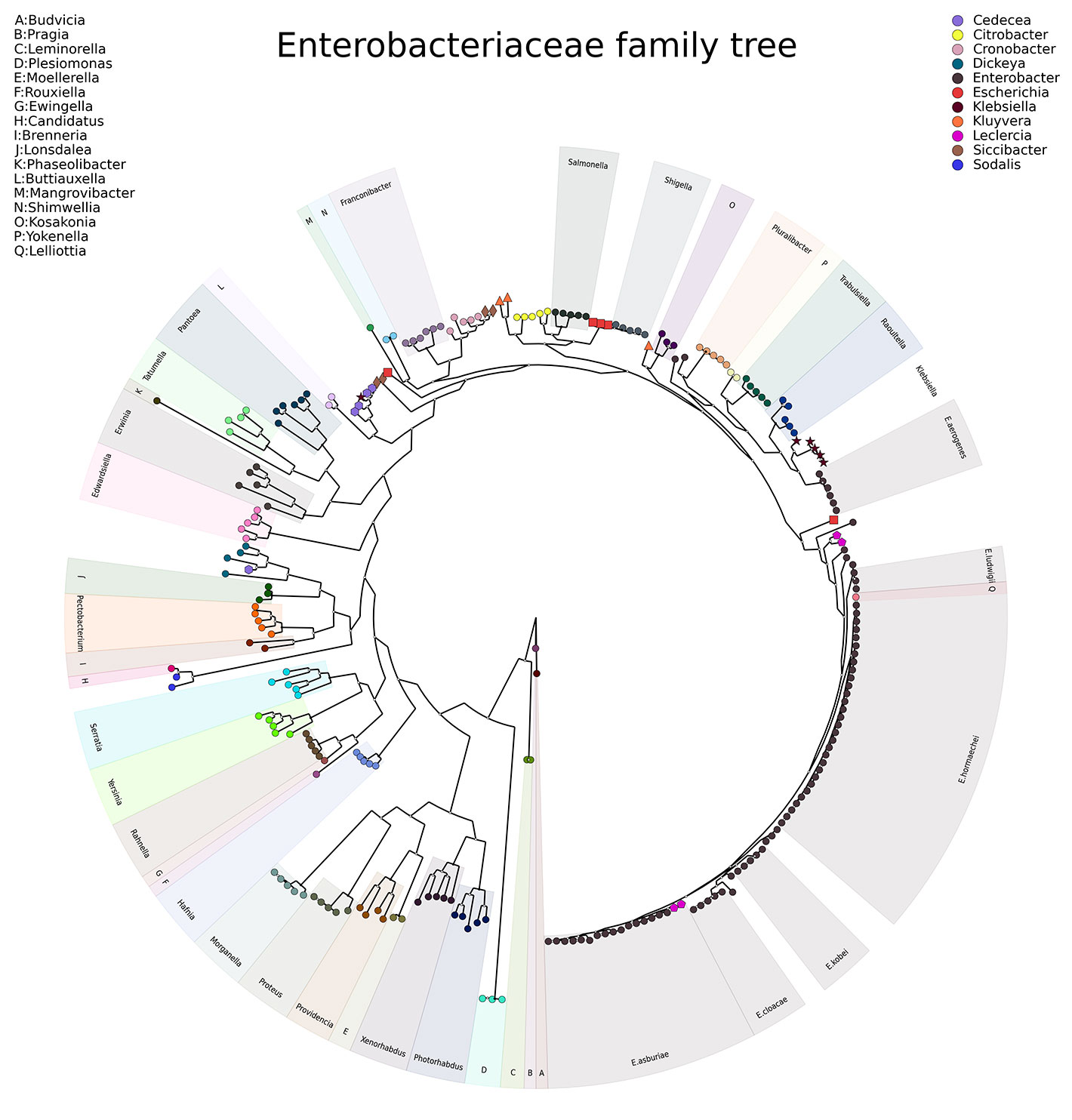
To investigate the relationship of Enterobacter aerogenes genomes in the context of other members of the family Enterobacteriaceae, LOCUST was used to construct a phylogenetic universal marker tree of representative genomes of members of the family Enterobacteriaceae. These results supported the previously proposed argument to rename E. aerogenes to Klebsiella aerogenes.
Publications
Frontiers in microbiology. 2017-09-06; 8.1661.
Strain Level Streptococcus Colonization Patterns during the First Year of Life
Bioinformatics (Oxford, England). 2017-06-01; 33.11: 1725-1726.
LOCUST: a custom sequence locus typer for classifying microbial isolates
mBio. 2016-12-13; 7.6:
Comprehensive Genome Analysis of Carbapenemase-Producing Enterobacter spp.: New Insights into Phylogeny, Population Structure, and Resistance Mechanisms
Genome biology. 2015-07-21; 16.1: 143.
A novel method of consensus pan-chromosome assembly and large-scale comparative analysis reveal the highly flexible pan-genome of Acinetobacter baumannii
Funding
This project has been funded in whole or part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under Award Number U19AI110819.
